Related publications: [HKN+06, HKN+08]
Fibroblast Growth Factors (FGF) are a family of proteins which play a key role in the process of cell signalling in a variety of contexts, for example wound healing. The mechanisms of the FGF signalling pathway are complex and not yet fully understood. In the case study we analyse a model of the pathway based on literature-derived information regarding the early stages of FGF signal propagation and which incorporates several features that have been reported to negatively regulate this propagation.
Our model incorporates protein-protein interactions (including competition for partners), phosphorylation and dephosphorylation, protein complex relocation and protein complex degradation (via ubiquitin-mediated proteolysis). The follow figure illustrates the different components in the pathway and their possible bindings.
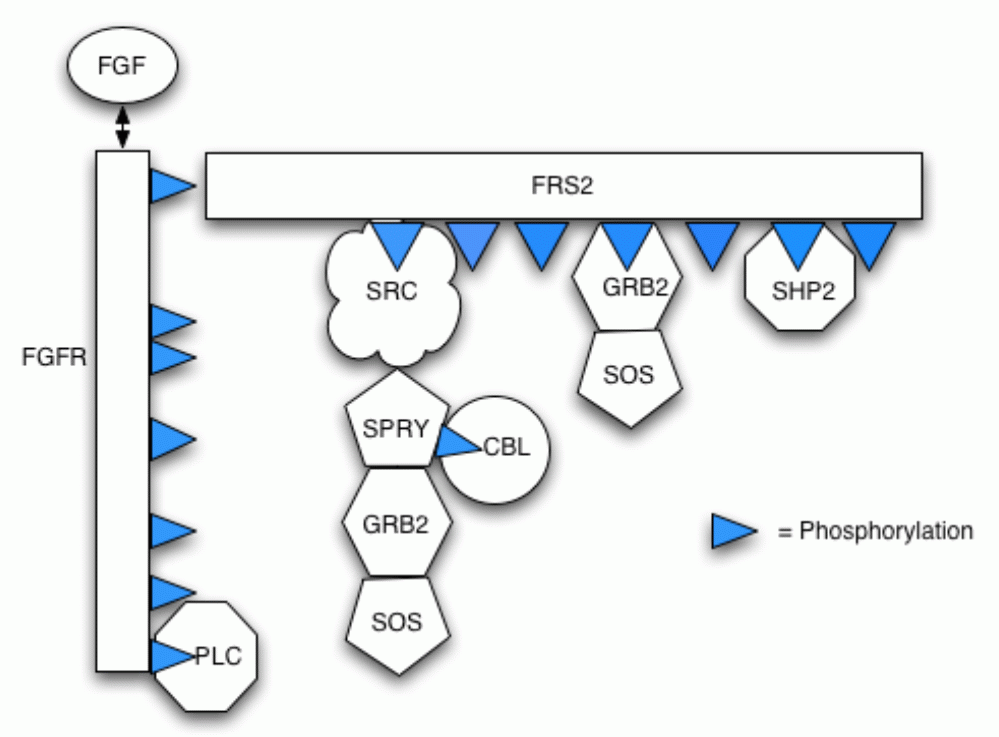
Below is a list of the reactions included in the model.
The full details of the reactions can be found here.
Note that this model is not intended to and cannot be a fully accurate representation of a real-world FGF signalling pathway. Its primary purpose at this stage of development is as a tool to evaluate biological hypotheses that are not easily obtained by intuition or manual methods. To this end, the model is an abstraction as argued in [RS02].
We explicitly draw attention to the following issues. The reactions selected are based upon their current biological interest rather than complete understanding of the components of FGF signalling. Indeed, at this stage we have ignored many reactions that could prove significant in regulation of FGFR signalling in real cells. However, the design permits the incorporation of further modifications to the core model as biological understanding advances. The model is idealised in that it does not take into account variations in composition, affinities or rate constants that might occur in different cell types or physiological conditions. However, a useful computational modelling approach should accommodate future quantitative or qualitative modifications to the core model, and we explicitly address this issue by evaluation of parameter dependencies below.
The model is based upon literature-defined events. There is always the possibility that the reported biological significance of these processes reflects the experimental context rather than the normal situation. For example, the significance of Plc in the relocation of FGFR signalling complexes has been the subject of some debate, as has the action of Spry. As we demonstrate in this paper, the model as presented provides the ability to ask what if questions which should inform the biological debate.
We now describe the specification in PRISM of the FGF model from the previous section. Each of the basic elements of the pathway, including all the possible compounds and receptors residues (FGF, FGFR, FRS2, Plc, Src, Spry, Sos, Grb2, Cbl and Shp2) is represented by a separate PRISM module and synchronisation between modules is used to model reactions which involve interactions between multiple elements. However, the different forms which each can take (for example, which other compounds it is bound to) are represented by one or more variables within the module. Our model represents a single instance of the signalling pathway, i.e. there can be at most one of each compound.
The study of a single instance of the pathway is also motivated by the fact that the same signal dynamics were obtained for a model where the concentrations were set to 100. Details of this model are available from here - note that this study uses the stochastic pi-calculus and the simulation tool BioSpi.
We constructed several models. In the first model, representing the full system, we suppose that initially FGF, unbound and unphosphorylated FGFR, unphosphorylated FRS2, unbound Src, Grb2, Cbl, Plc and Sos are all present in the system (Spry arrives into the system with the half-time of 10 minutes).
Subsequently, we constructed additional four models in order to investigate the roles of the various components of the activated receptor complex in controlling signalling dynamics. This is done by building modified models of the pathway where certain components are omitted (Shp2, Src, Spry or Plc). This is easily achieved in the PRISM model by just changing the initial value of the component under study.
The PRISM model for the pathway is given below.
// FGF model // gxn/dxp 10/02/06 // model is a CTMC ctmc // formulae formula frs = relocFRS2=0 & degFRS2=0; // frs2 not degraded or relocated formula fgfr = degFGFR=0; // fgfr not degraded or relocated //----------------------------------------------------------------------- // modules for the different components module FGF FGF : [0..1] init 1; // free // fgfr+fgf <-> fgfr:fgf [1] [fgf_bind] FGF=1 -> (FGF'=0); [fgf_rel] FGF=0 -> (FGF'=1); endmodule module FGFR FGFR : [0..1] init 1; // FGFR unbound to FRS2 degFGFR : [0..1] init 0; // FGFR degraded FGFR_FGF : [0..1] init 0; // FGF bound FGFR_PLC : [0..1] init 0; // PLC bound // phosporilation of receptors Y653P : [0..1] init 0; Y654P : [0..1] init 0; Y766P : [0..1] init 0; // fgfr+fgf <-> fgfr:fgf [1] [fgf_bind] fgfr & FGFR_FGF=0 -> 5000 : (FGFR_FGF'=1); [fgf_rel] fgfr & FGFR_FGF=1 -> 0.002 : (FGFR_FGF'=0); // phosporilation of receptors [] fgfr & FGFR_FGF=1 & Y653P=0 -> 0.1 : (Y653P'=1); // FGFR653 -> FGFR653P [2] [] fgfr & FGFR_FGF=1 & Y654P=0 -> 0.1 : (Y654P'=1); // FGFR654 -> FGFR654P [2] [] fgfr & Y653P=1 & Y654P=1 & Y766P=0 -> 70 : (Y766P'=1); // FGFR766 -> FGFR766P [3] // fgfr+frs2 <-> fgfr:frs2 [4] [fgfr_bind] fgfr & FGFR=1 -> (FGFR'=0); [fgfr_rel] fgfr & FGFR=0 -> (FGFR'=1); // PLC + FGFR766P <-> PLC:FGFR [9] [plc_bind] fgfr & Y766P=1 & FGFR_PLC=0 -> 10 : (FGFR_PLC'=1); [plc_rel] fgfr & FGFR_PLC=1 -> 0.02 : (FGFR_PLC'=0); // PLC:FGFR -> degFGFR [9] [] fgfr & FGFR_PLC=1 -> 1/(60*60) : (degFGFR'=1); endmodule module PLC PLC : [0..1] init 1; // free // PLC + FGFR766P <-> PLC:FGFR [9] [plc_bind] PLC=1 -> (PLC'=0); [plc_rel] PLC=0 -> (PLC'=1); endmodule module FRS2 relocFRS2 : [0..1] init 0; // ubiquitin mdification of FRS2 degFRS2 : [0..1] init 0; // FRS2 relocated FRS2_Ubi : [0..1] init 0; // FRS2 degraded // phosporilation of receptors Y196P : [0..1] init 0; Y306P : [0..1] init 0; Y471P : [0..1] init 0; // compounds:FRS2 FRS2_FGFR : [0..1] init 0; // 0 - FGFR not bound, 1 - FGFR bound FRS2_GRB : [0..2] init 0; // 0 - GRB2 not bound, 1 - GRB2 bound, 2 - GRB2:SOS bound FRS2_SHP : [0..1] init 0; // 0 - SHP2 not bound, 1 - SHP2 bound FRS2_SRC : [0..8] init 0; // 0 - SRC not bound // 1 - SRC bound // 2 - SRC:SPRY bound // 3 - SRC:SPRYP bound // 4 - SRC:SPRYP:CBL bound // 5 - SRC:SPRYP:GRB bound // 6 - SRC:SPRYP:GRB:CBL bound // 7 - SRC:SPRYP:GRB:SOS bound // 8 - SRC:SPRYP:GRB:SOS:CBL bound // fgfr+frs2 <-> fgfr:frs2 [4] [fgfr_bind] frs & FRS2_FGFR=0 -> 10 : (FRS2_FGFR'=1); [fgfr_rel] frs & FRS2_FGFR=1 -> 0.001 : (FRS2_FGFR'=0); // phosporilation of receptors [] frs & Y653P=1 & Y654P=1 & FRS2_FGFR=1 & Y196P=0 -> 0.2 : (Y196P'=1); // FRS2196 -> FRS2196P [5] [] frs & Y653P=1 & Y654P=1 & FRS2_FGFR=1 & Y306P=0 -> 0.2 : (Y306P'=1); // FRS2306 -> FRS2306P [5] [] frs & Y653P=1 & Y654P=1 & FRS2_FGFR=1 & Y471P=0 -> 0.2 : (Y471P'=1); // FRS2471 -> FRS2471P [5] // FRS2196 <- FRS2196P [6] [] frs & FRS2_SHP=1 & Y196P=1 & FRS2_SRC=0 -> 12 : (Y196P'=0); // src not bound [src_rel] frs & FRS2_SHP=1 & Y196P=1 & FRS2_SRC>0 -> 12 : (Y196P'=0) & (FRS2_SRC'=0); // src bound // FRS2306 <- FRS2306P [6] [] frs & FRS2_SHP=1 & Y306P=1 & FRS2_GRB=0 -> 12 : (Y306P'=0); // grb2 not bound [grb_rel] frs & FRS2_SHP=1 & Y306P=1 & FRS2_GRB>0 -> 12 : (Y306P'=0) & (FRS2_GRB'=0); // grb2 bound // FRS2471 <- FRS2471P [6] [shp_rel] frs & FRS2_SHP=1 & Y471P=1 -> 12 : (Y471P'=0) & (FRS2_SHP'=0); // SRC + FRS2196P <-> SRC:FRS2 [7] [src_bind] frs & Y196P=1 & FRS2_SRC=0 -> 10 : (FRS2_SRC'=SRC); [src_rel] frs & FRS2_SRC>0 -> 0.02 : (FRS2_SRC'=0); // GRB2 + FRS2306P <-> GRB2:FRS2 [7] [grb_bind] frs & Y306P=1 & FRS2_GRB=0 -> 10 : (FRS2_GRB'=GRB); [grb_rel] frs & FRS2_GRB>0 -> 0.02 : (FRS2_GRB'=0); // SHP2 + FRS2471P <-> SHP2:FRS2 [7] [shp_bind] frs & Y471P=1 & FRS2_SHP=0 -> 10 : (FRS2_SHP'=SHP); [shp_rel] frs & FRS2_SHP=1 -> 0.02 : (FRS2_SHP'=0); // Src:FRS2 -> degFRS2 [8] [] frs & FRS2_SRC>0 -> 1/(15*60) : (relocFRS2'=1); // Spry + Src -> Spry55:Src/Spryp + Src -> Spry55p:Src [11] [spry_bind_frs] frs & FRS2_SRC=1 -> 1 : (FRS2_SRC'=SPRY+1); // Spry + Src <- Spry55:Src [11] [spry_rel_frs] frs & FRS2_SRC=2 -> 0.01 : (FRS2_SRC'=1); // Spryp + Src <- Spry55p:Src [11] [spry_rel_frs] frs & FRS2_SRC>2 -> 0.0001 : (FRS2_SRC'=1); // Spry55:Src -> Spry55p:Src [11] [] frs & FRS2_SRC=2 -> 10 : (FRS2_SRC'=3); // Spry55p + Cbl <-> Spry55p:Cbl [11] [cbl_bind_frs] frs & (FRS2_SRC=3|FRS2_SRC=5|FRS2_SRC=7)-> 1 : (FRS2_SRC'=FRS2_SRC+1); [cbl_rel_frs] frs & (FRS2_SRC=4|FRS2_SRC=6|FRS2_SRC=8)-> 0.0001 : (FRS2_SRC'=FRS2_SRC-1); // Spry55p + Grb2 <-> Spry55p:Grb2 [11] [grb_bind_frs] frs & (FRS2_SRC=3|FRS2_SRC=4)-> 1 : (FRS2_SRC'=FRS2_SRC+2*GRB); [grb_rel_frs] frs & (FRS2_SRC=5|FRS2_SRC=6)-> 0.0001 : (FRS2_SRC'=FRS2_SRC-2); // not sos:grb2 [grb_rel_frs] frs & (FRS2_SRC=7|FRS2_SRC=8)-> 0.0001 : (FRS2_SRC'=FRS2_SRC-4); // sos:grb2 // Spry55p:Cbl +FRS2 -> Frs-Ubi [12] [] frs & (FRS2_SRC=4|FRS2_SRC=6|FRS2_SRC=8)& FRS2_Ubi=0 -> 0.00085 : (FRS2_Ubi'=1); // FRS2-Ubi -> degFRS2 [12] [] frs & FRS2_Ubi=1 -> 1/(5*60) : (degFRS2'=1); // Spry55p -> Spry55 [13] [spry_dephos] frs & FRS2_SHP=1 & FRS2_SRC>2 -> 12 : (FRS2_SRC'=2); // Grb2 + Sos <-> Grb2:Sos [14] [sos_bind_frs] frs & FRS2_GRB=1 -> 1 : (FRS2_GRB'=2);// grb2:frs2 [sos_rel_frs] frs & FRS2_GRB=2 -> 0.0001 : (FRS2_GRB'=1); // grb2:frs2 [sos_bind_frs] frs & (FRS2_SRC=5|FRS2_SRC=6)-> 1 : (FRS2_SRC'=FRS2_SRC+2); // grb2:spry [sos_rel_frs] frs & (FRS2_SRC=7|FRS2_SRC=8)-> 0.0001 : (FRS2_SRC'=FRS2_SRC-2); // grb2:spry endmodule module SRC SRC : [0..8] init 1; // 0 - bound elsewhere // 1 - SRC // 2 - SRC_SPRY // 3 - SRC_SPRYP // 4 - SRC_SPRYP_CBL // 5 - SRC_SPRYP_GRB // 6 - SRC_SPRYP_GRB_CBL // 7 - SRC_SPRYP_GRB_SOS // 8 - SRC_SPRYP_GRB_SOS_CBL // SRC + FRS2196P <-> SRC:FRS2 [7] [src_bind] SRC>0 -> (SRC'=0); [src_rel] SRC=0 -> (SRC'=FRS2_SRC); // Spry + Src -> Spry55:Src/Spryp + Src -> Spry55p:Src [11] [spry_bind] SRC=1 -> 1 : (SRC'=SPRY+1); // Spry + Src <- Spry55:Src [11] [spry_rel] SRC=2 -> 0.01 : (SRC'=1); // Spryp + Src <- Spry55p:Src [11] [spry_rel] SRC>2 -> 0.0001 : (SRC'=1); // Spry55:Src -> Spry55p:Src [11] [] SRC=2 -> 10 : (SRC'=3); // Spry55p + Cbl <-> Spry55p:Cbl [11] [cbl_bind_src] (SRC=3|SRC=5|SRC=7)-> 1 : (SRC'=SRC+1); [cbl_rel_src] (SRC=4|SRC=6|SRC=8)-> 0.0001 : (SRC'=SRC-1); // Spry55p + Grb2 <-> Spry55p:Grb2 [11] [grb_bind_src] (SRC=3|SRC=4)-> 1 : (SRC'=SRC+2*GRB); [grb_rel_src] (SRC=5|SRC=6)-> 0.0001 : (SRC'=SRC-2); // not sos:grb2 [grb_rel_src] (SRC=7|SRC=8)-> 0.0001 : (SRC'=SRC-4); // sos:grb2 // Grb2 + Sos <-> Grb2:Sos [14] [sos_bind_src] (SRC=5|SRC=6)-> 1 : (SRC'=SRC+2); [sos_rel_src] (SRC=7|SRC=8)-> 0.0001 : (SRC'=SRC-2); endmodule module SPRY SPRY : [0..7] init 0; // 0 - bound elsewhere or not appeared // 1 SPRY // 2 SPRYP // 3 SPRYP_CBL // 4 SPRYP_GRB // 5 SPRYP_GRB_CBL // 6 SPRYP_GRB_SOS // 7 SPRYP_GRB_SOS_CBL app : [0..1] init 0; // has spry entered the system yet // Spry + Src <-> Spry55:Src/Spryp + Src -> Spry55p:Src [11] [spry_bind] SPRY>0 -> (SPRY'=0); // src free [spry_bind_frs] SPRY>0 -> (SPRY'=0); // src:frs2 [spry_rel] SPRY=0 & SRC>0 -> (SPRY'=SRC-1); // src free [spry_rel_frs] SPRY=0 & FRS2_SRC>0 -> (SPRY'=FRS2_SRC-1); // src:frs2 // -> Spry [10] [] SPRY=0 & app=0 -> 1/(15*60) : (SPRY'=1) & (app'=1); // Spry55p + Cbl <-> Spry55p:Cbl [11] [cbl_bind] (SPRY=2|SPRY=4|SPRY=6)-> 1 : (SPRY'= SPRY+1); [cbl_rel] (SPRY=3|SPRY=5|SPRY=7)-> 0.0001 : (SPRY'= SPRY-1); // Spry55p + Grb2 <-> Spry55p:Grb2 [11] [grb_bind_spry] (SPRY=2|SPRY=3)-> 1 : (SPRY'= SPRY+2*GRB); [grb_rel_spry] (SPRY=4|SPRY=5)-> 0.0001 : (SPRY'= SPRY-2); // not sos:grb2 [grb_rel_spry] (SPRY=6|SPRY=7)-> 0.0001 : (SPRY'= SPRY-4); // sos:grb2 // Grb2 + Sos <-> Grb2:Sos [14] [sos_bind_spry] (SPRY=4|SPRY=5)-> 1 : (SPRY'=SPRY+2); [sos_rel_spry] (SPRY=6|SPRY=7)-> 0.0001 : (SPRY'=SPRY-2); endmodule module CBL CBL : [0..1] init 1; // free // Spry55p + Cbl <-> Spry55p:Cbl [11] [cbl_bind] CBL=1 -> (CBL'=0); // spryp free [cbl_bind_src] CBL=1 -> (CBL'=0); // spryp:src and not src:frs2 [cbl_bind_frs] CBL=1 -> (CBL'=0); // spryp:src and src:frs2 [cbl_rel] CBL=0 -> (CBL'=1); // spryp free [cbl_rel_src] CBL=0 -> (CBL'=1); // spryp:src and not src:frs2 [cbl_rel_frs] CBL=0 -> (CBL'=1); // spryp:src and src:frs2 // Spry55p -> Spry55 [13] [spry_dephos] true -> (CBL'=1); endmodule module SHP SHP : [0..1] init 1; // free // SHP2 + FRS2471P <-> SHP2:FRS2 [7] [shp_bind] SHP=1 -> (SHP'=0); [shp_rel] SHP=0 -> (SHP'=1); endmodule module GRB GRB : [0..2] init 1; // 1 free and 2 grb2:sos // Grb2 + Sos <-> Grb2:Sos [14] [sos_bind] GRB=1 -> 1 : (GRB'=2); [sos_rel] GRB=2 -> 0.0001 : (GRB'=1); // GRB2 + FRS2306P <-> GRB2:FRS2 [7] [grb_bind] GRB>0 -> (GRB'=0); [grb_rel] GRB=0 -> (GRB'=FRS2_GRB); // Spry55p + Grb2 <-> Spry55p:Grb2 [11] [grb_bind_spry] GRB>0 -> (GRB'=0); // spryp free [grb_bind_src] GRB>0 -> (GRB'=0); // spryp:src and not src:frs2 [grb_bind_frs] GRB>0 -> (GRB'=0); // spryp:src and src:frs2 [grb_rel_spry] GRB=0 -> (GRB'=(SPRY<6)?1:2); // spryp free [grb_rel_src] GRB=0 -> (GRB'=(SRC<7)?1:2); // spryp:src and not src:frs2 [grb_rel_frs] GRB=0 -> (GRB'=(FRS2_SRC<7)?1:2); // spryp:src and src:frs2 // Spry55p -> Spry55 [13] [spry_dephos] true -> (GRB'=(FRS2_SRC<7)?1:2); endmodule module SOS SOS : [0..1] init 1; // Grb2 + Sos <-> Grb2:Sos [14] [sos_bind] SOS=1 -> (SOS'=0); // grb2 free [sos_bind_spry] SOS=1 -> (SOS'=0); // grb2:spryp and not spryp:src [sos_bind_src] SOS=1 -> (SOS'=0); // grb2:spryp and spryp:src and not src:frs2 [sos_bind_frs] SOS=1 -> (SOS'=0); // grb2:spryp and spryp:src and src:frs2 [sos_rel] SOS=0 -> (SOS'=1); // grb2 free [sos_rel_spry] SOS=0 -> (SOS'=1); // grb2:spryp and not spryp:src [sos_rel_src] SOS=0 -> (SOS'=1); // grb2:spryp and spryp:src and not src:frs2 [sos_rel_frs] SOS=0 -> (SOS'=1); // grb2:spryp and spryp:src and src:frs2 endmodule // reward structure: number of bindings rewards "bindings" [grb_bind] degFGFR=0 & relocFRS2=0 & degFRS2=0 : 1; endrewards // reward structure: time bound rewards "bound" FRS2_GRB>0 & relocFRS2=0 & degFRS2=0 : 1; endrewards // reward structure: time rewards "time" true : 60; endrewards
The table below shows statistics for the MTBDD which represents each model we have built, in terms of the number of states, transitions, the total number of nodes and the number of leaves (terminal nodes).
| model | States | Transitions | Nodes | Leaves | |
| full model | 80,616 | 560,520 | 26,122 | 17 | |
| no Shp2 | 14,088 | 91,752 | 9,607 | 16 | |
| no Src | 1,200 | 7,848 | 4,107 | 14 | |
| no Spry | 1,176 | 6,928 | 6,766 | 14 | |
| no Plc | 40,440 | 276,544 | 22,784 | 16 |
The table below gives the times taken to construct the models. There are two stages. Firstly, the system description (in our module based language) is parsed and converted to the MTBDD representing it. Secondly, reachability is performed to identify non-reachable states and the MTBDD is filtered accordingly. Reachability is performed using a BDD based fixpoint algorithm. The number of iterations for this process is also given.
| model | Time (s) | Iter.s | |
| full model | 0.51 | 32 | |
| no Shp2 | 0.17 | 25 | |
| no Src | 0.15 | 17 | |
| no Spry | 0.17 | 18 | |
| no Plc | 0.43 | 30 |
Our primary goal in this case study is to analyse the various mechanisms previously reported to negatively regulate signalling. Since the binding of Grb2 to FRS2 serves as the primary link between FGFR activation and ERK signalling, we examine the amount of Grb2 bound to FRS2 as the system evolves. In addition we investigate the different causes of degradation which, from the system specification, can be caused by one of the following reactions occurring:
Below, we present a list of the various properties of the model that we have analysed, and the form in which they are specified to the PRISM tool.
These property can be expressed by the CSL by the following formulae:
const double T; // time bound // the probability that Grb2 is bound to FRS2 at the time instant T P=? [ F[T,T] FRS2_GRB>0 & relocFRS2=0 & degFRS2=0 ] // the expected number of times that Grb2 binds to FRS2 by time T R{"bindings"}=? [ C<=T ] // the expected time that Grb2 spends bound to FRS2 within the first T time units R{"bound"}=? [ C<=T ] // the long-run probability that Grb2 is bound to FRS2 S=? [ FRS2_GRB>0 & relocFRS2=0 & degFRS2=0 ] // the expected number of times Grb2 binds to FRS2 before either degradation or relocation occurs R{"bindings"}=? [ F relocFRS2=1 | degFRS2=1 | degFGFR=1 ] // the expected time Grb2 spends bound to FRS2 before either degradation or relocation occurs R{"bound"}=? [ F relocFRS2=1 | degFRS2=1 | degFGFR=1 ] // the probability that each possible cause of degradation/relocation has occurred by time T P=? [ relocFRS2=0 & degFRS2=0 & degFGFR=0 U[0,T] relocFRS2=1 ] P=? [ relocFRS2=0 & degFRS2=0 & degFGFR=0 U[0,T] degFRS2=1 ] P=? [ relocFRS2=0 & degFRS2=0 & degFGFR=0 U[0,T] degFGFR=1 ] // the probability that each possible cause of degradation/relocation occurs first P=? [ relocFRS2=0 & degFRS2=0 & degFGFR=0 U relocFRS2=1 ] P=? [ relocFRS2=0 & degFRS2=0 & degFGFR=0 U degFRS2=1 ] P=? [ relocFRS2=0 & degFRS2=0 & degFGFR=0 U degFGFR=1 ] // the expected time until either degradation or relocation occurs in the pathway R{"time"}=? [ F relocFRS2=1 | degFRS2=1 | degFGFR=1 ]
The probability that Grb2 is bound to FRS2 at the time instant T. In the graph below, we have plotted this measure as T varies from 0 to 60 minutes.
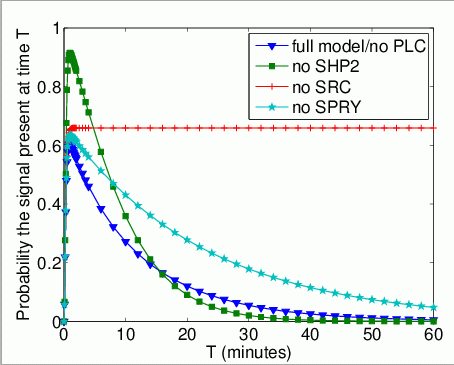
The expected number of times that Grb2 binds to FRS2 by time T.
In the graph below, we have plotted this measure as T varies from 0 to 60 minutes.
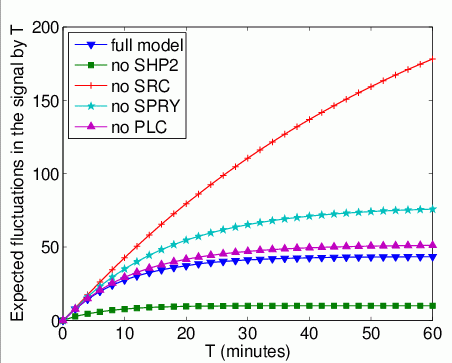
The expected time that Grb2 spends bound to FRS2 within the first T time units. In the graph below, we have plotted this measure as T varies from 0 to 60 minutes.
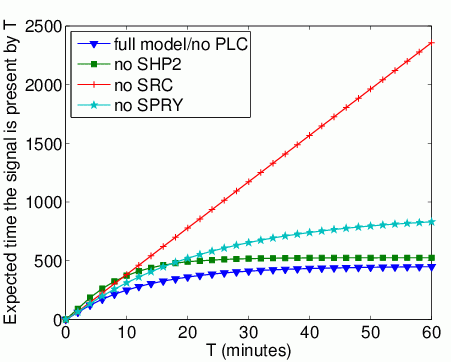
The long-run probability that Grb2 is bound to FRS2. In the table below we present the results obtained for this property.
| model | probability | |
| full model | 7.54e-7 | |
| no Shp2 | 3.29e-9 | |
| no Src | 0.659460 | |
| no Spry | 4.6e-6 | |
| no Plc | 0.0 |
The expected number of times Grb2 binds to FRS2 before either degradation or relocation occurs. In the table below we present the model check statistics and results obtained for this property.
| model | Iterations | Time per iter (s) | reward | ||
| full model | 5,934 | 0.019873 | 43.1027 | ||
| no Shp2 | 548 | 0.003365 | 10.0510 | ||
| no Src | 26,768 | 0.000169 | 283.233 | ||
| no Spry | 8,111 | 0.000235 | 78.3314 | ||
| no Plc | 6,828 | 0.008778 | 51.5475 |
The expected time Grb2 spends bound to FRS2 before either degradation or relocation occurs. In the table below we present the model check statistics and results obtained for this property.
| model | Iterations | Time per iter (s) | reward | ||
| full model | 5,937 | 0.025217 | 6.27042 | ||
| no Shp2 | 549 | 0.003973 | 7.78927 | ||
| no Src | 26,768 | 0.000201 | 39.6102 | ||
| no Spry | 8,110 | 0.000273 | 10.8791 | ||
| no Plc | 6,831 | 0.011287 | 7.56241 |
The probability that each possible cause of degradation/relocation has occurred by time T. In the graphs below, we have plotted these measures as T varies from 0 to 60 minutes.
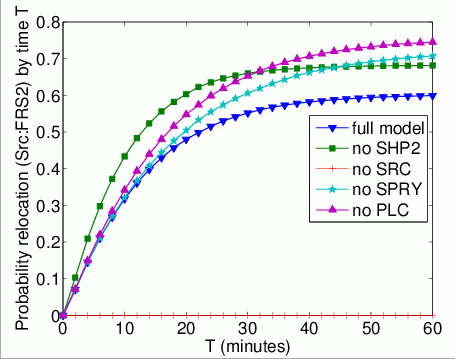
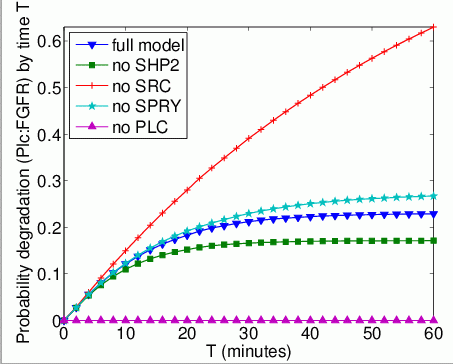
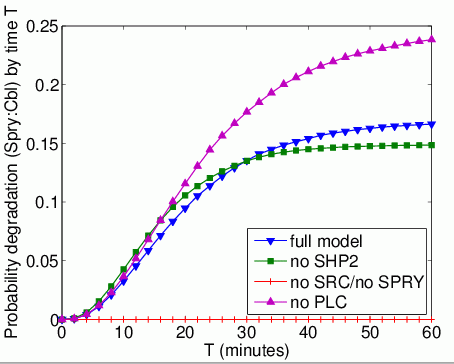
The probability that each possible cause of degradation/relocation occurs first. In the tables below we present the model check statistics and results obtained for these properties.
| model | Src:FRS2 | |||
| iters | Time per iter (s) | result: | ||
| full model | 5,965 | 0.017676 | 0.602356 | |
| no Shp2 | 583 | 0.002642 | 0.679102 | |
| no Src | - | - | - | |
| no Spry | 8,110 | 0.000155 | 0.724590 | |
| no Plc | 6,863 | 0.012363 | 0.756113 | |
| model | Plc:FGFR | |||
| iters | Time per iter (s) | result: | ||
| full model | 5,965 | 0.015472 | 0.229107 | |
| no Shp2 | 583 | 0.002521 | 0.176693 | |
| no Src | - | 1.0 | ||
| no Spry | 8,110 | 0.000155 | 0.275410 | |
| no Plc | - | - | - | |
| model | Spry:Cbl | |||
| iters | Time per iter (s) | result: | ||
| full model | 6,228 | 0.014881 | 0.168536 | |
| no Shp2 | 610 | 0.002951 | 0.149742 | |
| no Src | - | - | 0.0 | |
| no Spry | - | - | - | |
| no Plc | 7,124 | 0.011837 | 0.243887 | |
The expected time until either degradation or relocation occurs in the pathway. In the table below we present the model check statistics and results obtained for this property.
| model | Iterations | Time per iter (s) | reward (mins) | ||
| full model | 5,964 | 0.020113 | 14.0258 | ||
| no Shp2 | 583 | 0.003310 | 10.5418 | ||
| no Src | 26,767 | 0.000169 | 60.3719 | ||
| no Spry | 8,110 | 0.000231 | 16.8096 | ||
| no Plc | 6,863 | 0.008808 | 17.5277 |